This lab was created by Guillaume Bourque.
Table of contents
- Introduction
- Calling Variants with GATK
- Investigating the SNP calls
- Filter the variants
- Adding functional consequence
- Investigating the functional consequence of variants
- Adding dbSNP annotations
- Overall script
- (Optional) Investigating the trio
- Acknowledgements
Introduction
In one of the previous modules, we have aligned the reads from NA12878 (daughter) in a small region on chromosome 1. In this module, we will use the data in the BAM files to call variants and then we’ll perform some annotation and filtration.
As a brief reminder, these are the IDs for each member in the trio:
| Mother | Father | Child |
|---|---|---|
| NA12892 | NA12891 | NA12878 |
Our read were extracted from the following regions:
| Chromosome | Start | End |
|---|---|---|
| chr1 | 17704860 | 18004860 |
Preliminaries
Amazon node
Read these directions for information on how to log in to your assigned Amazon node.
Work directory
Create a new directory that will store all of the files created in this lab.
rm -rf ~/workspace/module5
mkdir -p ~/workspace/module5
cd ~/workspace/module5
ln -s ~/CourseData/HT_data/Module5/* .
Note:
The ln -s command adds symbolic links of all of the files contained in the (read-only) ~/CourseData/HT_data/Module5 directory.
Input files
Our starting data set consists of 100 bp paired-end Illumina reads from the child (NA12878) that have been aligned to hg19 during one of the previous modules (NA12878.bwa.sort.bam). We also have the same data after duplicate removal and realignment around indels NA12878.bwa.sort.rmdup.realign.bam.
If you type ls, you should have something like:
ubuntu@ip-10-182-231-187:~/workspace/module5$ ls
NA12878.bwa.sort.bam NA12878.bwa.sort.rmdup.realign.bai other_files
NA12878.bwa.sort.bam.bai NA12878.bwa.sort.rmdup.realign.bam
other_files is a directory that contains additional reference files and scripts that we will need during the module.
Do you know what are the .bai files?
If you’re interested, you can also look at the fastQC reports for the original fastq files in the directory:
~workspace/module5/other_files/fastQC_reports
You can view these reports at http://cbw##.dyndns.info/module5/other_files/fastQC_reports/.
Note: You need to replace ## by your student number.
Calling variants with GATK
If you recall from the previous module, we first mapped the reads to hg19 and then we removed duplicate reads and realigned the reads around the indels.
Let’s call SNPs in NA12878 using both the original and the improved bam files:
java -Xmx2g -jar /usr/local/GATK/GenomeAnalysisTK.jar -T UnifiedGenotyper -l INFO \
-R other_files/hg19.fa -I NA12878.bwa.sort.bam -stand_call_conf 30 -stand_emit_conf 10 \
-o NA12878.bwa.sort.bam.vcf -nt 4 -glm BOTH -L chr1:17704860-18004860
java -Xmx2g -jar /usr/local/GATK/GenomeAnalysisTK.jar -T UnifiedGenotyper -l INFO \
-R other_files/hg19.fa -I NA12878.bwa.sort.rmdup.realign.bam -stand_call_conf 30 \
-stand_emit_conf 10 -o NA12878.bwa.sort.rmdup.realign.bam.vcf -nt 4 \
-glm BOTH -L chr1:17704860-18004860
-Xmx2g instructs java to allow up 2 GB of RAM to be used for GATK.
-l INFO specifies the minimum level of logging.
-R specifies which reference sequence to use.
-I specifies the input BAM files.
-stand_call_conf is the minimum phred-scaled Qscore threshold to separate high confidence from low confidence calls (that aren’t at ‘trigger’ sites). Only genotypes with confidence >= this threshold are emitted as called sites. A reasonable threshold is 30 (this is the default).
-stand_emit_conf is the minimum phred-scaled Qscore threshold to emit low confidence calls (that aren’t at ‘trigger’ sites). Genotypes with confidence >= this but less than the calling threshold are emitted but marked as filtered. The default value is 30.
-L indicates the reference region where SNP calling should take place
-nt 4 specifies that GATK should use 4 processors.
-glm BOTH instructs GATK that we want to use both the SNP and INDEL genotype likelihoods models
File check
At this point, you should have the following result files
ubuntu@ip-10-182-231-187:~/workspace/module5$ ls
NA12878.bwa.sort.bam NA12878.bwa.sort.bam.vcf.idx NA12878.bwa.sort.rmdup.realign.bam.vcf
NA12878.bwa.sort.bam.bai NA12878.bwa.sort.rmdup.realign.bai NA12878.bwa.sort.rmdup.realign.bam.vcf.idx
NA12878.bwa.sort.bam.vcf NA12878.bwa.sort.rmdup.realign.bam other_files
Investigating the SNP calls
Use less to take a look at the vcf files:
less NA12878.bwa.sort.bam.vcf
less NA12878.bwa.sort.rmdup.realign.bam.vcf
A bit hard to read… Better with the -S option?
vcf is a daunting format at first glance, but you can find some basic information about the format here or here.
How do you figure out what the genotype is for each variant?
Do we have any annotation information yet?
How many SNPs were found?
Did we find the same number of variants using the files before and after duplicate removal and realignment?
Looking for differences between the two vcf files
Use the following command to pull out differences between the two files:
diff <(grep ^chr NA12878.bwa.sort.bam.vcf | cut -f1-2 | sort) \
<(grep ^chr NA12878.bwa.sort.rmdup.realign.bam.vcf | cut -f1-2 | sort)
Use IGV to investigate the SNPs
The best way to see and understand the differences between the two vcf files will be to look at them in IGV.
If you need, the IGV color codes can be found here: IGV color code
Option 1: You can view your files (bam and vcf files) in the IGV browser by using the URL for that file from your Cloud instance. We have a web server running on the Amazon cloud for each instance.
In a browser, like Firefox, type in your server name (cbw#.dyndns.info) and all files under your workspace will be shown there. Find your bam and your vcf files, right click it and ‘copy the link location’.
Next, open IGV and select hg19 as the reference genome as you did in the visualization module.
In IGV, load both the original and the realigned bam files (NA12878.bwa.sort.bam and NA12878.bwa.sort.rmdup.realign.bam) using (File->Load from URL…).
After you have loaded the two bam files, load the two vcf files (NA12878.bwa.sort.bam.vcf and NA12878.bwa.sort.rmdup.realign.bam.vcf) in the same way.
Option 2: Alternatively, you can download all the NA12878.* files in the current directory to your local computer:
NA12878.bwa.sort.bam NA12878.bwa.sort.bam.vcf.idx NA12878.bwa.sort.rmdup.realign.bam.vcf
NA12878.bwa.sort.bam.bai NA12878.bwa.sort.rmdup.realign.bai NA12878.bwa.sort.rmdup.realign.bam.vcf.idx
NA12878.bwa.sort.bam.vcf NA12878.bwa.sort.rmdup.realign.bam
To do this you can use the procedure that was described previously.
After that you need to follow the steps as in Option 1 except that you need to load the files in IGV using (File->Load from File…).
Finally, go to a region on chromsome 1 with reads and spend some time SNP gazing…
Do the SNPs look believable?
Are there any positions that you think should have been called as a SNP, but weren’t?
Did you find positions where the two files lead to different SNP calls?
Go back to some of the positions that were listed as different from the previous section, for instance:
chr1 17791668
You should see something like:
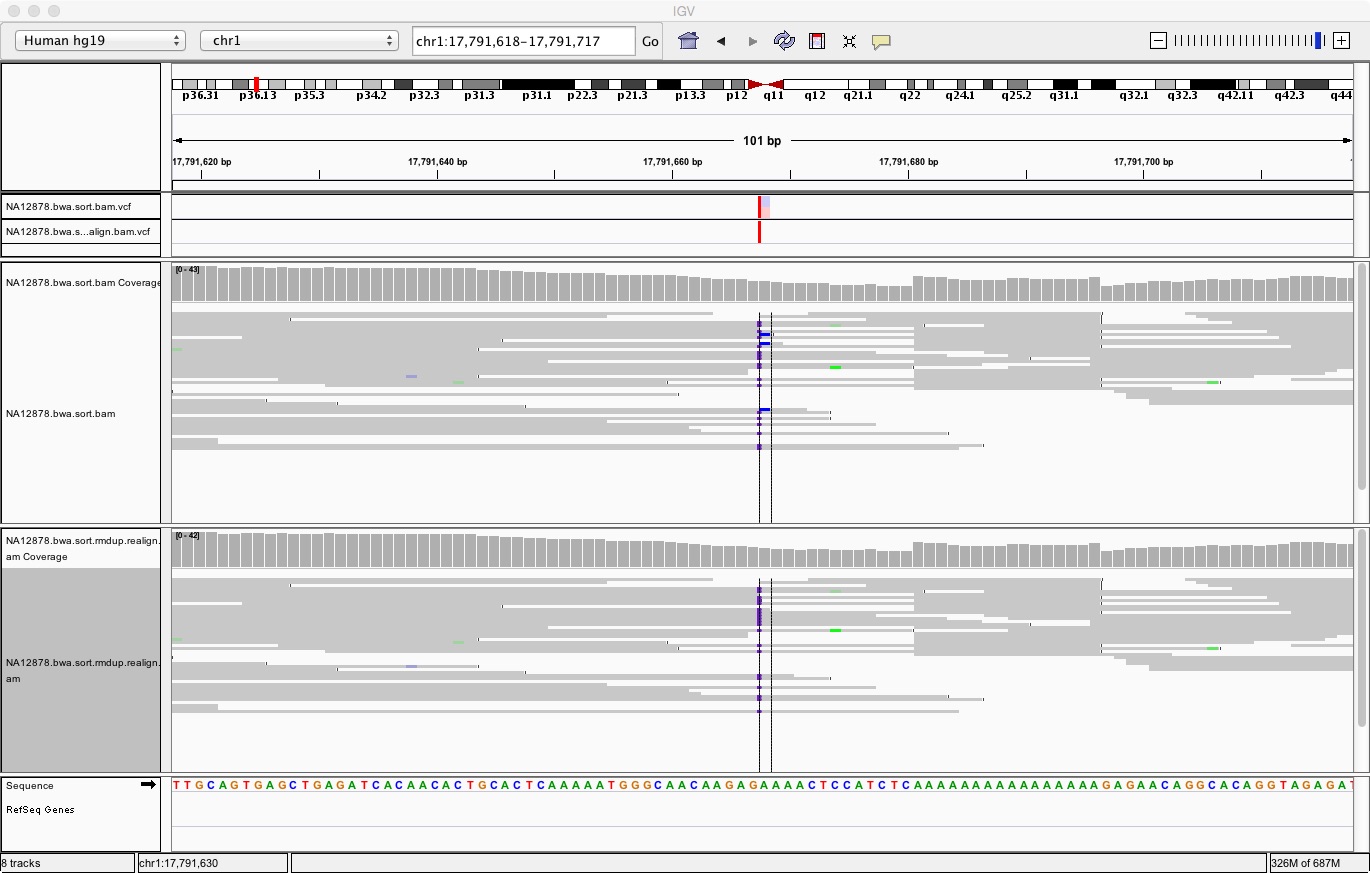
In the rest of this module we will focus on the SNPs calls found after duplicate removal and realignment.
Looking for INDELs
INDELs can be found by looking for rows where the reference base column and the alternate base column are different lengths. It’s slightly more complicated than that since, you’ll also pick up the comma delimited alternate bases.
Here’s an awk expression that almost picks out the INDELs:
grep -v "^#" NA12878.bwa.sort.rmdup.realign.bam.vcf \
| awk '{ if(length($4) != length($5)) { print $0 } }' \
| less -S
You can find a slightly more advanced awk script that separates the SNPs from the INDELs here.
Did you find any INDELs?
Can you find the largest INDEL?
Filter the variants
Typically variant callers will only perform a minimal amount of filtering when presenting variant calls. In the case of GATK, we are actively removing any variant with score less than 10. Any variant with a score less than 30 is labeled with the filter “LowQual”.
To perform more rigorous filtering, another program must be used. In our case, we will use the VariantFiltration tool in GATK.
NOTE: The best practice when using GATK is to use the VariantRecalibrator. In our data set, we had too few variants to accurately use the variant recalibrator and therefore we used the VariantFiltration tool instead.
java -Xmx2g -jar /usr/local/GATK/GenomeAnalysisTK.jar -T VariantFiltration \
-R other_files/hg19.fa --variant NA12878.bwa.sort.rmdup.realign.bam.vcf \
-o NA12878.bwa.sort.rmdup.realign.bam.filter.vcf --filterExpression "QD < 2.0" \
--filterExpression "FS > 200.0" --filterExpression "MQ < 40.0" \
--filterName QDFilter --filterName FSFilter --filterName MQFilter
-Xmx2g instructs java to allow up 2 GB of RAM to be used for GATK.
-R specifies which reference sequence to use.
--variant specifies the input vcf file.
-o specifies the output vcf file.
--filterExpression defines an expression using the vcf INFO and genotype variables.
--filterName defines what the filter field should display if that filter is true.
What is QD, FS, and MQ?
File check
At this point, you should have the following result files:
ubuntu@ip-10-182-231-187:~/workspace/module5$ ls
NA12878.bwa.sort.bam NA12878.bwa.sort.rmdup.realign.bai NA12878.bwa.sort.rmdup.realign.bam.vcf
NA12878.bwa.sort.bam.bai NA12878.bwa.sort.rmdup.realign.bam NA12878.bwa.sort.rmdup.realign.bam.vcf.idx
NA12878.bwa.sort.bam.vcf NA12878.bwa.sort.rmdup.realign.bam.filter.vcf other_files
NA12878.bwa.sort.bam.vcf.idx NA12878.bwa.sort.rmdup.realign.bam.filter.vcf.idx
Adding functional consequence
The next step in trying to make sense of the variant calls is to assign functional consequence to each variant.
At the most basic level, this involves using gene annotations to determine if variants are sense, missense, or nonsense.
java -Xmx2G -jar other_files/snpEff/snpEff.jar eff \
-c other_files/snpEff/snpEff.config -v -no-intergenic \
-i vcf -o vcf hg19 NA12878.bwa.sort.rmdup.realign.bam.filter.vcf \
> NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.vcf
-Xmx2g instructs java to allow up 4 GB of RAM to be used for snpEff.
-c specifies the path to the snpEff configuration file
-v specifies verbose output.
-no-intergenic specifies that we want to skip functional consequence testing in intergenic regions.
-i and -o specify the input and output file format respectively. In this case, we specify vcf for both.
hg19 specifies that we want to use the hg19 annotation database.
NA12878.bwa.sort.rmdup.realign.bam.filter.vcf specifies our input vcf filename
NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.vcf specifies our output vcf filename
File check
At this point, you should have the following files:
ubuntu@ip-10-182-231-187:~/workspace/module5$ ls
NA12878.bwa.sort.bam NA12878.bwa.sort.rmdup.realign.bam.filter.vcf
NA12878.bwa.sort.bam.bai NA12878.bwa.sort.rmdup.realign.bam.filter.vcf.idx
NA12878.bwa.sort.bam.vcf NA12878.bwa.sort.rmdup.realign.bam.vcf
NA12878.bwa.sort.bam.vcf.idx NA12878.bwa.sort.rmdup.realign.bam.vcf.idx
NA12878.bwa.sort.rmdup.realign.bai other_files
NA12878.bwa.sort.rmdup.realign.bam snpEff_genes.txt
NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.vcf snpEff_summary.html
Investigating the functional consequence of variants
You can learn more about the meaning of snpEff annotations here.
Use less to look at the new vcf file:
less NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.vcf
The annotation is presented in the INFO field using the new ANN format. For more information on this field see here. Typically, we have:
ANN=Allele|Annotation|Putative impact|Gene name|Gene ID|Feature type|Feature ID|Transcript biotype|Rank / Total|HGVS.c|...
Here’s an example of a typical annotation:
ANN=C|intron_variant|MODIFIER|PADI6|PADI6|transcript|NM_207421.4|Coding|5/16|c.553+80T>C||||||
What does the example annotation actually mean?
Next, you should view or download the report generated by snpEff.
Use the procedure described previously to retrieve:
snpEff_summary.html
Next, open the file in any web browser.
Finding impactful variants
One nice feature in snpEff is that it tries to assess the impact of each variant. You can read more about the effect categories here.
Let’s begin by looking for variants with a high impact:
grep HIGH NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.vcf
How many variants had a high impact? What effect categories were represented in these variants?
Open that position in IGV, what do you see?
Let’s continue by looking for variants with a moderate impact:
grep MODERATE NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.vcf
How many variants had a moderate impact? What effect categories were represented in these variants?
Adding dbSNP annotations
Go back to looking at your last vcf file:
less -S NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.vcf
What do you see in the third column?
The third column in the vcf file is reserved for identifiers. Perhaps the most common identifier is the dbSNP rsID.
Use the following command to generate dbSNP rsIDs for our vcf file:
java -Xmx2g -jar /usr/local/GATK/GenomeAnalysisTK.jar -T VariantAnnotator -R other_files/hg19.fa \
--dbsnp other_files/dbSNP_135_chr1.vcf.gz --variant NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.vcf \
-o NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.dbsnp.vcf -L chr1:17704860-18004860
-Xmx2g instructs java to allow up 2 GB of RAM to be used for GATK.
-R specifies which reference sequence to use.
--dbsnp specifies the input dbSNP vcf file. This is used as the source for the annotations.
--variant specifies the input vcf file.
-o specifies the output vcf file.
-L defines which regions we should annotate. In this case, I chose the chromosomes that contain the regions we are investigating.
What percentage of the variants that passed all filters were also in dbSNP?
grep -v ^# NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.dbsnp.vcf | grep PASS | grep -c rs
grep -v ^# NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.dbsnp.vcf | grep -c PASS
Can you find a variant that wasn’t in dbSNP?
File Check
At this point, you should have the following files:
ubuntu@ip-10-182-231-187:~/workspace/module5$ ls
NA12878.bwa.sort.bam NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.vcf.idx
NA12878.bwa.sort.bam.bai NA12878.bwa.sort.rmdup.realign.bam.filter.vcf
NA12878.bwa.sort.bam.vcf NA12878.bwa.sort.rmdup.realign.bam.filter.vcf.idx
NA12878.bwa.sort.bam.vcf.idx NA12878.bwa.sort.rmdup.realign.bam.vcf
NA12878.bwa.sort.rmdup.realign.bai NA12878.bwa.sort.rmdup.realign.bam.vcf.idx
NA12878.bwa.sort.rmdup.realign.bam other_files
NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.dbsnp.vcf snpEff_genes.txt
NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.dbsnp.vcf.idx snpEff_summary.html
NA12878.bwa.sort.rmdup.realign.bam.filter.snpeff.vcf
Overall script
NOTE: If you ever become truly lost in this lab, you can use the lab script to automatically perform all of the steps listed here. If you are logged into your CBW account, just run: ~/CourseData/HT_data/Module5/other_files/RunModule5.sh. You can also download the file if you want to bring it home with you.
(Optional) Investigating the trio
At this point we have aligned and called variants in one individual. However, we actually have FASTQ and BAM files for three family members!
As additional practice, perform the same steps for the other two individuals (her parents): NA12891 and NA12892. Here are some additional things that you might want to look at:
-
If you load up all three realigned BAM files and all three final vcf files into IGV, do the variants look plausible? Use a Punnett square to help evaluate this. i.e. if both parents have a homozygous reference call and the child has a homozygous variant call at that locus, this might indicate a trio conflict.
-
Do you find any additional high or moderate impact variants in either of the parents?
-
Do all three family members have the same genotype for Rs7538876 and Rs2254135?
-
GATK produces even better variant calling results if all three BAM files are specified at the same time (i.e. specifying multiple
-I filenameoptions). Try this and then perform the rest of module 5 on the trio vcf file. Does this seem to improve your variant calling results? Does it seem to reduce the trio conflict rate?
Acknowledgements
This module is heavily based on a previous module prepared by Michael Stromberg.