Every year after the workshops, we make a public Amazon server image (AMI) based on the server we used. This image contains all the tools we used in the workshop.
Launching the AMI
- Sign up for an AWS account
- You want to launch a computing server – EC2

- Click the launch button
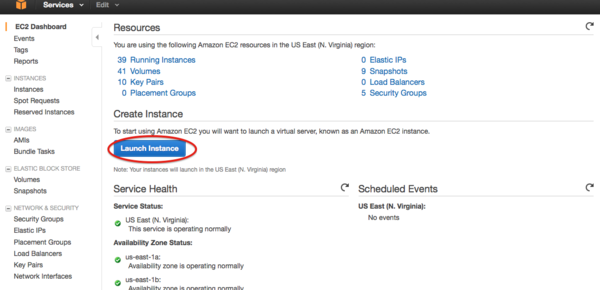
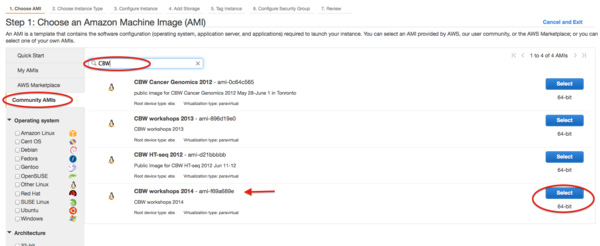
- Select CBW image by searching “Community AMIs” with “CBW” or AMI id and choose the one you used in the workshop.
- Choose an instance type, we used m3.xlarge in the workshop
- Configure Instance (you can use the default)
- Add storage for workspace
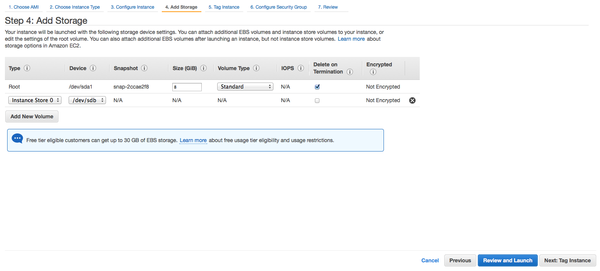
- Add tags if you want
- Configure security group to allow services like ssh, http, …
- You need to create or choose key pair for the new server. Remember to download the key pair you created or you will not be able connect to the new server.
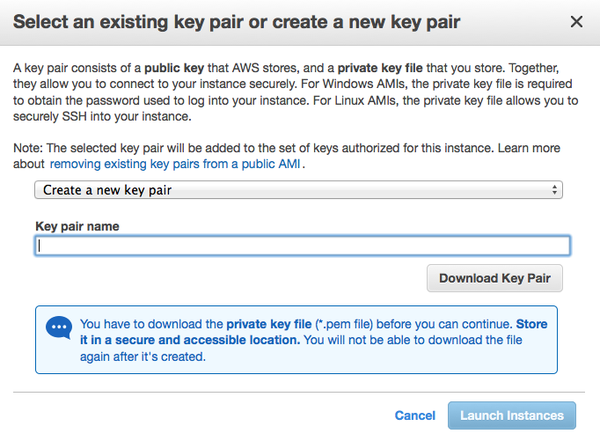
- Launch!
Data set
Please follow the links on our wiki page to download the data sets we used in the workshop. You may need load public data sets that contains the reference genomes (e.g. hg19). You can upload your own or download from some sources like we talked about in the workshop. Free available S3 buckets are 1000 Genomes, GenBank, etc. http://aws.amazon.com/datasets?_encoding=UTF8&jiveRedirect=1