This lab was created by Guillaume Bourque.
Table of contents
- Overview
- Align DNA with BWA-MEM
- Characterize the fragment size distribution
- Run DELLY to detect SVs
- Setting up IGV for SV visualization
- Explore the SVs
- Overall script
- (Optional) Look for other SVs
- Acknowledgements
Overview
The goal of this practical session is to identify structural variants (SVs) in a human genome by identifying both discordant paired-end alignments and split-read alignments that. If you recall from the lecture, discordant paired-end alignments conflict with the alignment patterns that we expect (i.e., concordant alignments) for the DNA library and sequencing technology we have used. For example, given a ~500bp paired-end Illumina library, we expect pairs to align in F/R orientation and we expect the ends of the pair to align roughly 500bp apart. Pairs that align too far apart suggest a potential deletion in the DNA sample’s genome. As you may have guessed, the trick is how we define what “too far” is — this depends on the fragment size distribution of the data. Split-read alignments contain SV breakpoints and consequently, then DNA sequences up- and down-stream of the breakpoint align to disjoint locations in the reference genome.
In this session, we will use DELLY, a SV detection tool. DELLY is an integrated structural variant prediction method that can discover, genotype and visualize deletions, tandem duplications, inversions and translocations at single-nucleotide resolution in short-read massively parallel sequencing data. It uses paired-ends and split-reads to sensitively and accurately delineate genomic rearrangements throughout the genome. If you are interested in DELLY, you can read the full manuscript here.
The dataset we are using comes from the Illumina Platinum Genomes Project, which is a 50X-coverage dataset of the NA12891/NA12892/NA12878 trio. The raw data can be downloaded from the following URL.
Our focus will be on chromosome 20, as processing three whole human genomes worth of data is intractable given the time allowed for this session.
Preliminaries
Amazon node
Read these directions for information on how to log in to your assigned Amazon node.
Work directory
Let’s create a new work directory for this module within our workspace:
rm -rf ~/workspace/module6
mkdir -p ~/workspace/module6
cd ~/workspace/module6
Create symbolic links to all of the files that are relevant to the SV discovery exercise.
ln -s ~/CourseData/HT_data/Module6/* .
Note:
The ln -s command adds symbolic links of all of the files contained in the (read-only) ~/CourseData/HT_data/Module6 directory.
In this step, we created a new directory that will store all of the files created in this lab. The ln -s command adds symbolic links of all of the files contained in the ~/CourseData sub-directories that are relevant to the SV module. This directory contains all of the alignment files (in BAM format, as you will recall) and genome annotations that we’ll need during the lab.
At this point you should have the following files:
ubuntu@ip-10-164-192-186:~/workspace/module6$ ls
bam delly_call fastq pairend_distro.py reference RunModule6.sh
Looking in the bam directory, you should see:
ubuntu@ip-10-164-192-186:~/workspace/module6$ ls bam
backup NA12878_S1.chr20.20X.pairs.posSorted.bam.bai
discordants NA12891_S1.chr20.20X.pairs.posSorted.bam
NA12878.molelculo.chr20.bam NA12891_S1.chr20.20X.pairs.posSorted.bam.bai
NA12878.molelculo.chr20.bam.bai NA12892_S1.chr20.20X.pairs.posSorted.bam
NA12878.pacbio.chr20.bam NA12892_S1.chr20.20X.pairs.posSorted.bam.bai
NA12878.pacbio.chr20.bam.bai README.md
NA12878_S1.chr20.20X.pairs.posSorted.bam validated
Align DNA with BWA-MEM
This step has been done for you in the interest of time, but the commands are shown so that you can reproduce the results later. The advantage of using BWA-MEM in the context of SV discovery is that it produces both paired-end and split-read alignments in a single BAM output file. In contrast, prior to BWA-MEM, one typically had to use two different aligners in order to produce both high quality paired-end and split-read alignments.
In the alignment commands, note the use of the -M parameter to mark shorter split hits as secondary.
#################
# Align NA12878 #
#################
# bwa mem hg19.fa \
# fastq/NA12878_S1.chr20.20X.1.fq \
# fastq/NA12878_S1.chr20.20X.2.fq \
# -M \
# | samtools view -S -b - \
# > bam/backup/NA12878_S1.chr20.20X.pairs.readSorted.bam
#################
# Align NA12891 #
#################
# bwa mem hg19.fa \
# fastq/NA12891_S1.chr20.20X.1.fq \
# fastq/NA12891_S1.chr20.20X.2.fq \
# -M \
# | samtools view -S -b - \
# > bam/backup/NA12891_S1.chr20.20X.pairs.readSorted.bam
#################
# Align NA12892 #
#################
# bwa mem hg19.fa \
# fastq/NA12892_S1.chr20.20X.1.fq \
# fastq/NA12892_S1.chr20.20X.2.fq \
# -M \
# | samtools view -S -b - \
# > bam/backup/NA12892_S1.chr20.20X.pairs.readSorted.bam
Characterize the fragment size distribution
We have used BWA-MEM to align all of the Illumina paired-end sequences (in FASTQ format) to the human genome. Before we can attempt to identify structural variants via discordant alignments, we must first characterize the fragment size distribution — this describes the size of concordant (i.e., they align in the expected orientation and with the expected distance between the ends) alignments, and the corollary is that we can also use the size distribution to decide the size threshold for discordant alignments. The following script, taken from the distribution of LUMPY extracts F/R pairs from a BAM file and computes the mean and stdev of the F/R alignments. It also generates a density plot of the fragment size distribution.
Calculation of the fragment distribution for NA12878.
samtools view bam/backup/NA12878_S1.chr20.20X.pairs.readSorted.bam \
| ./pairend_distro.py -r 101 -X 4 -N 10000 \
-o NA12878_S1.chr20.20X.pairs.histo \
> NA12878_S1.chr20.20X.pairs.params
Calculation of the fragment distribution for NA12891.
samtools view bam/backup/NA12891_S1.chr20.20X.pairs.readSorted.bam \
| ./pairend_distro.py -r 101 -X 4 -N 10000 \
-o NA12891_S1.chr20.20X.pairs.histo \
> NA12891_S1.chr20.20X.pairs.params
Calculation of the fragment distribution for NA12892.
samtools view bam/backup/NA12892_S1.chr20.20X.pairs.readSorted.bam \
| ./pairend_distro.py -r 101 -X 4 -N 10000 \
-o NA12892_S1.chr20.20X.pairs.histo \
> NA12892_S1.chr20.20X.pairs.params
At this point you should have the following files:
ubuntu@ip-10-164-192-186:~/workspace/module6$ ls
bam NA12878_S1.chr20.20X.pairs.histo NA12891_S1.chr20.20X.pairs.params pairend_distro.py
delly_call NA12878_S1.chr20.20X.pairs.params NA12892_S1.chr20.20X.pairs.histo reference
fastq NA12891_S1.chr20.20X.pairs.histo NA12892_S1.chr20.20X.pairs.params RunModule6.sh
Let’s take a peak at the first few lines of the histogram file that was produced:
head -n 10 NA12878_S1.chr20.20X.pairs.histo
Expected results:
0 0.0
1 0.000200461060439
2 0.000300691590659
3 0.000300691590659
4 0.000200461060439
5 0.000200461060439
6 0.000300691590659
7 0.00010023053022
8 0.00010023053022
9 0.00010023053022
Let’s use R to plot the fragment size distribution. First, launch R from the command line.
R
Now, within R, execute the following commands:
size_dist <- read.table('NA12878_S1.chr20.20X.pairs.histo')
pdf(file = "NA12878.fragment.hist.pdf")
plot(size_dist[,1], size_dist[,2], type='h')
dev.off()
size_dist <- read.table('NA12891_S1.chr20.20X.pairs.histo')
pdf(file = "NA12891.fragment.hist.pdf")
plot(size_dist[,1], size_dist[,2], type='h')
dev.off()
size_dist <- read.table('NA12892_S1.chr20.20X.pairs.histo')
pdf(file = "NA12892.fragment.hist.pdf")
plot(size_dist[,1], size_dist[,2], type='h')
dev.off()
quit("no")
At this point, you should have the following files:
ubuntu@ip-10-164-192-186:~/workspace/module6$ ls
bam NA12878_S1.chr20.20X.pairs.params NA12892_S1.chr20.20X.pairs.histo
delly_call NA12891.fragment.hist.pdf NA12892_S1.chr20.20X.pairs.params
fastq NA12891_S1.chr20.20X.pairs.histo pairend_distro.py
NA12878.fragment.hist.pdf NA12891_S1.chr20.20X.pairs.params reference
NA12878_S1.chr20.20X.pairs.histo NA12892.fragment.hist.pdf RunModule6.sh
If successful, you should be able to access the 3 PDF files at the following URLs:
http://cbwXX.dyndns.info/module6/NA12878.fragment.hist.pdf
http://cbwXX.dyndns.info/module6/NA12891.fragment.hist.pdf
http://cbwXX.dyndns.info/module6/NA12892.fragment.hist.pdf
Note: Copy and paste the URL below into your browser, but change the “XX” in the URL to your student number.
Spend some time thinking about what this plot means for identifying discordant alignments.
What does the mean fragment size appear to be?
Are all 3 graphs the same?
Is that good or bad?
Run DELLY to detect SVs
For germline SVs, calling is done by sample or in small batches to increase SV sensitivity & breakpoint precision. At this point we will only call deletions. Here are the steps adapted from the DELLY readme.
Call deletions
Let’s start with NA12878:
delly call -t DEL -g reference/hg19.fa -o NA12878.bcf -x reference/hg19.excl \
bam/NA12878_S1.chr20.20X.pairs.posSorted.bam
.bcf files are compressed vcf files. To look at the output you can use:
bcftools view NA12878.bcf | less -S
Now NA12891 and NA12892:
delly call -t DEL -g reference/hg19.fa -o NA12891.bcf -x reference/hg19.excl \
bam/NA12891_S1.chr20.20X.pairs.posSorted.bam
delly call -t DEL -g reference/hg19.fa -o NA12892.bcf -x reference/hg19.excl \
bam/NA12892_S1.chr20.20X.pairs.posSorted.bam
Cheat: If these commands are taking too long, simply run the command cp delly_call/* .
Merge calls
We need to merge the SV sites into a unified site list:
delly merge -t DEL -m 500 -n 1000000 -o del.bcf -b 500 -r 0.5 \
NA12878.bcf NA12891.bcf NA12891.bcf
Re-genotype in all samples
We need to re-genotype the merged SV site list across all samples. This can be run in parallel for each sample.
delly call -t DEL -g reference/hg19.fa -v del.bcf -o NA12878.geno.bcf \
-x reference/hg19.excl bam/NA12878_S1.chr20.20X.pairs.posSorted.bam
delly call -t DEL -g reference/hg19.fa -v del.bcf -o NA12891.geno.bcf \
-x reference/hg19.excl bam/NA12891_S1.chr20.20X.pairs.posSorted.bam
delly call -t DEL -g reference/hg19.fa -v del.bcf -o NA12892.geno.bcf \
-x reference/hg19.excl bam/NA12892_S1.chr20.20X.pairs.posSorted.bam
Merge the new calls
Merge all re-genotyped samples to get a single VCF/BCF using bcftools merge. Also index the resulting file and create vcf file for visualization.
bcftools merge -O b -o merged.bcf NA12878.geno.bcf NA12891.geno.bcf NA12892.geno.bcf
bcftools index merged.bcf
bcftools view merged.bcf > merged.vcf
Apply a filter for germline events
delly filter -t DEL -f germline -o germline.bcf -g reference/hg19.fa merged.bcf
bcftools view germline.bcf > germline.vcf
Do you know how to look at the resulting file?
File check
At this point you should have the following files:
ubuntu@ip-10-164-192-186:~/workspace/module6$ ls
bam NA12878.bcf.csi NA12891_S1.chr20.20X.pairs.params
del.bcf NA12878.fragment.hist.pdf NA12892.bcf
del.bcf.csi NA12878.geno.bcf NA12892.bcf.csi
delly_call NA12878.geno.bcf.csi NA12892.fragment.hist.pdf
fastq NA12878_S1.chr20.20X.pairs.histo NA12892.geno.bcf
germline.bcf NA12878_S1.chr20.20X.pairs.params NA12892.geno.bcf.csi
germline.bcf.csi NA12891.bcf NA12892_S1.chr20.20X.pairs.histo
germline.vcf NA12891.bcf.csi NA12892_S1.chr20.20X.pairs.params
merged.bcf NA12891.fragment.hist.pdf pairend_distro.py
merged.bcf.csi NA12891.geno.bcf reference
merged.vcf NA12891.geno.bcf.csi RunModule6.sh
NA12878.bcf NA12891_S1.chr20.20X.pairs.histo
Setting up IGV for SV visualization
Launch IGV and load the merged calls and the germline calls using File -> Load from URL using:
http://cbwXX.dyndns.info/module6/merged.vcf
http://cbwXX.dyndns.info/module6/germline.vcf
Note: Once again you will need to replace XX by your student number.
Navigate to the following location to see a deletion:
chr20:31,308,410-31,315,294
Now load the bam files in the same way using:
http://cbwXX.dyndns.info/module6/bam/NA12878_S1.chr20.20X.pairs.posSorted.bam
http://cbwXX.dyndns.info/module6/bam/NA12891_S1.chr20.20X.pairs.posSorted.bam
http://cbwXX.dyndns.info/module6/bam/NA12892_S1.chr20.20X.pairs.posSorted.bam
You should see something like this:
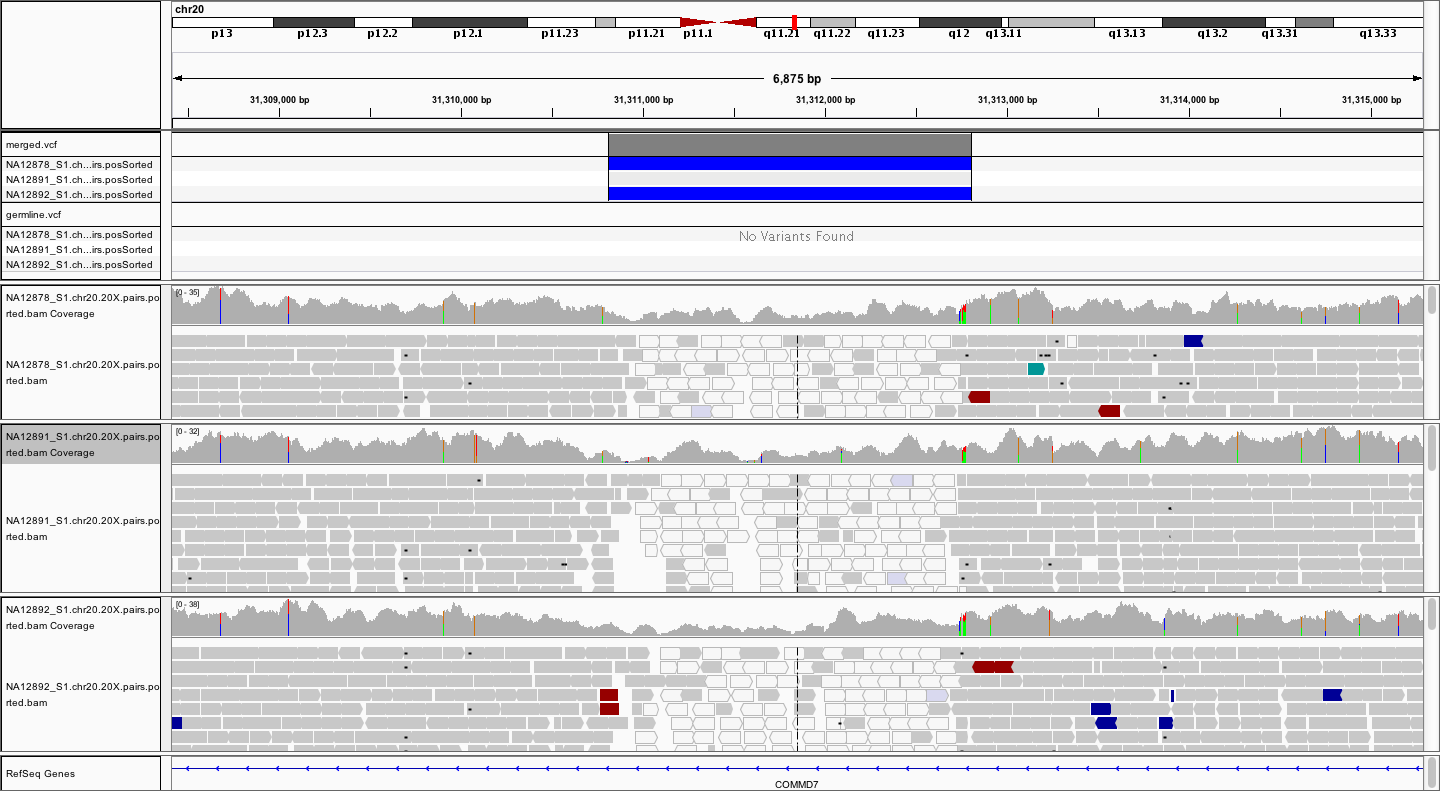
You can try to configure IGV such that we can more clearly see the alignments that support the SV prediction.
Do you remember how to make sure that the alignments are colored by insert size and orientation?
To further validate that there is a deletion there you can also another track which was generate using the Moleculo long read technology:
http://cbwXX.dyndns.info/module6/bam/NA12878.molelculo.chr20.bam
Explore the SVs
Is the variant at chr20:31,310,769-31,312,959 found in each member of the trio?
What are the genotypes for each member of the trio at this locus (e.g., hemizygous, homozygous)?
What about the variant at chr20:37,054,372-37,056,562?
Does the evidence in the Moleculo track mimic the evidence in the Illumina track for NA12878?
What about chr20:42,269,896-42,278,072?
Continue exploring the data!
Overall script
NOTE: If you ever become truly lost in this lab, you can use the lab script to automatically perform all of the steps listed here. If you are logged into your CBW account, just run: ~/CourseData/HT_data/Module6/RunModule6.sh. You can also download the file if you want to bring it home with you.
(Optional) Look for other SVs
You can try using Delly to look for other types of SVs, for example using:
delly -t DUP
delly -t INV
delly -t TRA
Acknowledgements
This module is heavily based on a previous module prepared by Aaron Quinlan.